Developing nuclear gene markers
Cross-species target gene enrichment
Data filtering in phylogenomic analysis
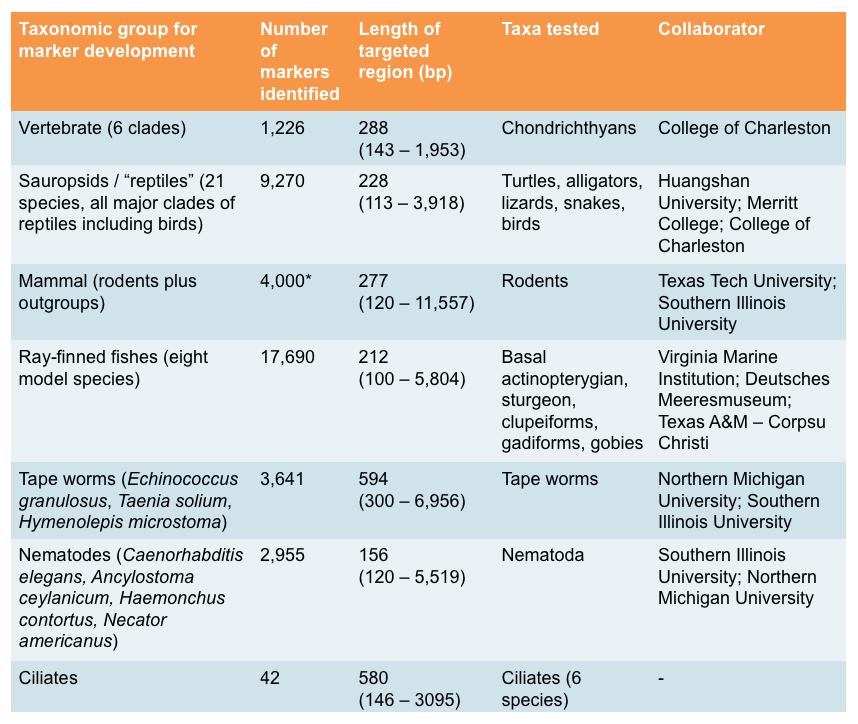
Tabel 1. Target gene markers have been developed and tested in our lab
EvolMarkers is a database based on genome comparison to find conserved single-copy exon (CDS) and intron (EPIC) markers for phylogenetic and population studies.
The cost of high-throughput sequencing is decreasing rapidly, from which we benefit as genome sequence data become more and more affordable. Sequencing the whole genome, however, is still too expensive and oftentimes not necessary for phylogenetic and population genetic studies. Target gene capture is a good alternative tool for those studies, because it could be used to collect sequence data of only the interested regions and to gather data from many individual samples in a single sequencing run when samples are multiplexed. We have developed strategies and protocols in our lab in the last few years for target gene capture and applied those in phylogenetics and population genomics, and a few money saving tricks used in library preparation and gene capture.
The use of genome-scale data to infer phylogenetic relationships has surged in recent years due to progress made in target-gene capture methods and sequencing techniques. Data filtering, the approach of excluding undesirable data from analyses, presumably could alleviate problems caused by systematic errors in phylogenetic inference. Data filtering approaches are now more relevant than ever as there are often thousands of loci available in genome-scale data, allowing for a more rigorous data selection scheme. Different data filtering criteria, such as evolutionary rate, base composition, phylogenetic informativeness, stemminess, and molecular clock-likeness as well as others have been proposed for selecting useful phylogenetic markers. Nevertheless, there are still few studies testing all these criteria simultaneously. We tested the separate and joint effects of those data filtering criteria. We found through carefully examining different characteristics of molecular markers, that calibrated clock-likeness is the best indicator of the phylogenetic usefulness of molecular markers in our study. Stemminess, the ratio of internal branch lengths over the total tree length is not a good indicator of phylogenetic performance. Slow-evolving genes are also not necessarily strong prior candidates for phylogenetic analysis. Phylogenetic informativeness can be informative when trying to resolve specific nodes in a tree, such as those associated with short internal branches, but may not be a good indicator of the tree-wide usefulness of a locus. Base compositional bias, as indicated by relative composition variability (RCV) values, may be a useful indicator of problematic phylogenetic markers, particularly by employing a threshold cutoff to exclude outliers with very high RCV values.
Environment DNA (eDNA) is free DNA from organisms living in the environment. It is thought that eDNA release into water or soil though damaged skin, feces, mucus of the organisms, and etc. As early as in 2008, eDNA has been used to identify freshwater invertebrates Ficetola et al., 2008). Currently, eDNA is broadly applied in evaluation and conservation of fish and other aquatic animals (Foote et al. 2012, Thomsen et al. 2012, Kelly et al. 2014a, Miya et al. 2015). Comparing to traditional biological survey, eDNA is more sensitive, cost efficient, and impose little or no damage to the environment (Thomsen and Willerslev 2015), so that it become one of the research line with greatest potential in recent years. However, there are two bottlenecks limiting the growth of eDNA approach: 1) inefficiency in the current PCR based eDNA approach; 2) no good quantification method using eDNA data. We are developing new eDNA methods based on gene capture and a novel quantification strategy using eDNA data.